Schedule > Posters
Espaces posters et démos
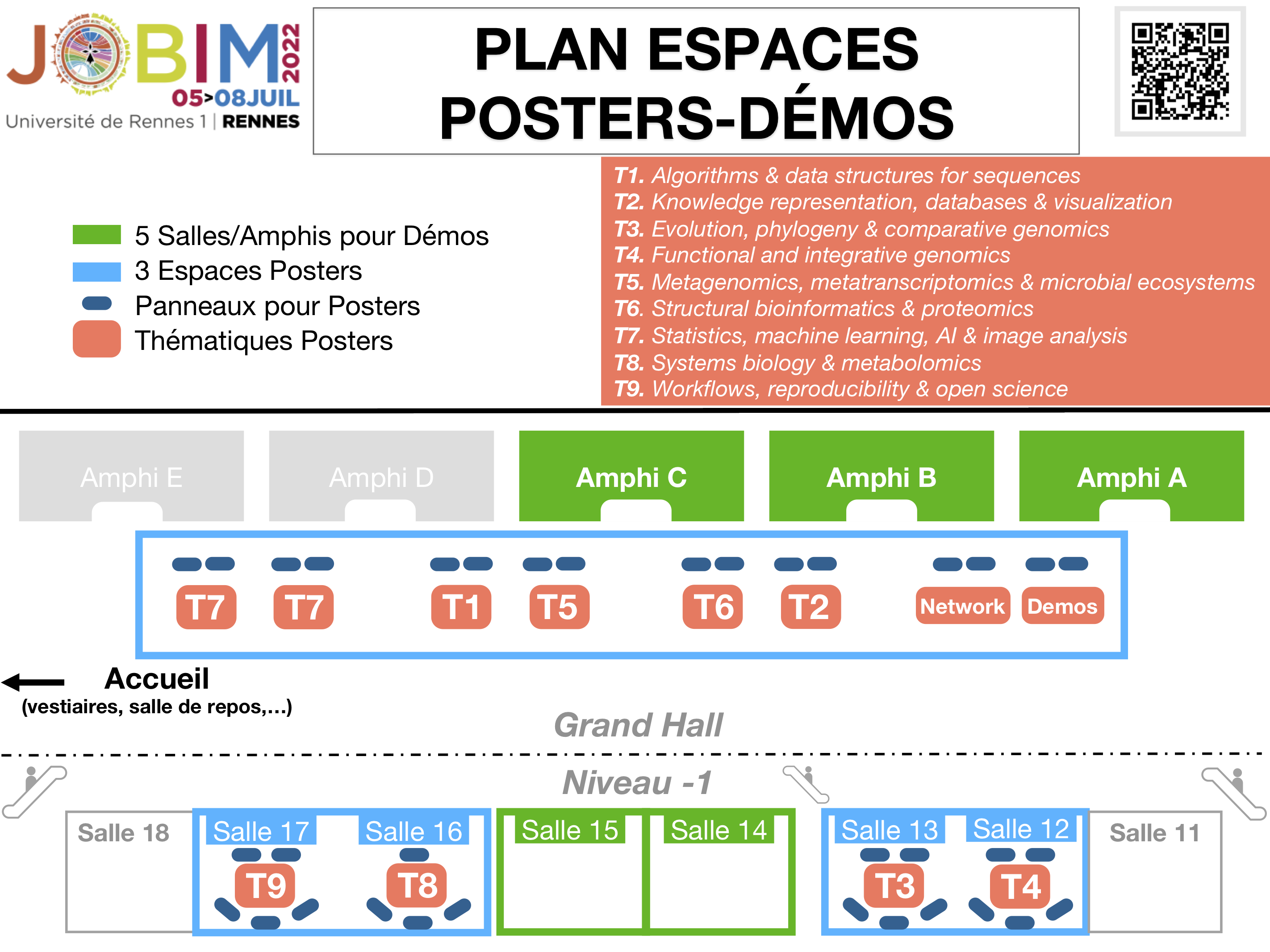
Poster list
The mapping between the easychair submission identifiers and the new number by topics is available in this pdf file.
| T1: Algorithms & data structures for sequences |
|---|
Wed. 6th |
| T1.2 |
ROCK: digital normalization of whole genome sequencing data (Alexis Criscuolo) |
| T1.4 |
Évaluation comparative des méthodes d’alignement multiple de séquences appliquées au séquençage de troisième génération (Coralie Rohmer) |
| T1.6 |
SVJedi-graph: genotyping close and overlapping structural variants with a variation graph and long-reads (Sandra Romain) |
| T1.8 |
A novel weight-based approach to determine cell type specificity from single cell datasets (Yanis Habtoun) |
| T1.10 |
Memory efficient subsampling strategy for large scale analysis of sequencing data (Timothé Rouzé) |
| T1.12 |
Interpreting mass spectra differing from their peptide models by several modifications (Albane Lysiak) |
| T1.14 |
ToulligQC 2: fast and comprehensive quality control for Oxford Nanopore sequencing data (Laurent Jourdren) |
| T1.16 |
A nextflow workflow for peptide sequence design in a targeted proteomic approach (Sylvère Bastien) |
Thu. 7th |
| T1.1 |
EstiAge: A tool to estimate the age of a variant (Thomas Ludwig) |
| T1.3 |
msscaf: a multiple source genome scaffolder (Matthias Zytnicki) |
| T1.5 |
Preliminary Results from Nanopore Q20+ sequencing (Claire Kuchly) |
| T1.7 |
Breakpoint detection in shallow and targeted panel in Rhabdomyosarcomas (Camille Benoist) |
| T1.9 |
Identifying Copy Number Variations in exome data – applications to infertility. (Amandine Septier) |
| T1.11 |
AptaMat: a matrix-based algorithm to compare single-stranded oligonucleotides secondary structures (Thomas Binet) |
| T1.13 |
MAM : Methylation Analysis of Microalgae. (Simon Brocard) |
| T1.15 |
Benchmark of Nvidia Clara Parabricks suite for GPU-accelerated bioinformatic processing of raw RNA-seq data (Etienne Bardet) |
| T2: Knowledge representation, databases & visualization |
|---|
Wed. 6th |
| T2.2 |
Improving attribute exploration for the detection and correction of anomalies in an agroecological knowledge base (Nassif Saab) |
| T2.4 |
The Phaeoexplorer Database: a Multi-Scale Genomic and Transcriptomic Data Resource for the Brown Algae (Loraine Brillet-Guéguen) |
| T2.6 |
Omnicrobe, an open-access database of microbial habitats, phenotypes and uses extracted from text (Sandra Dérozier) |
| T2.8 |
The advantage of having a web dedicated group in a Bioinformatic team (Rachel Torchet) |
| T2.10 |
Heterogeneous biological data integration with Semantic Web technologies using RDF or RDF-Star formalisms generate topologically different graphs (Christophe Heligon) |
| T2.12 |
APPINetwork : an R package for building and computational analysis of protein-protein interaction networks (Simon Gosset) |
| T2.14 |
Computational analysis of molecules, olfactory receptors, odors and their interactions (Guillaume Ollitrault) |
| T2.16 |
In-silico genome-wide detection of common fragile sites in human cells using BrdU-seq data (Nathan Alary) |
Thu. 7th |
| T2.1 |
MetExploreViz: Visualization tool for metabolic networks (Jean-Clément Gallardo) |
| T2.3 |
MyGOD : Interface de visualisation et d’analyse de données provenant des observatoires génomiques marins (Charlotte André) |
| T2.5 |
Accessibilité des modèles pharmacocinétiques physiologiques (Sidonie Halluin) |
| T2.7 |
IDy-Path : Identification Dynamique des épidémies de Pathogènes (Bryce Leterrier) |
| T2.9 |
Cirscan: a shiny application to decipher circRNA-miRNA-mRNA networks from multi-level transcriptomic data (Rose-Marie Fraboulet) |
| T2.11 |
Revisiting iPPI-DB as a tool to navigate the pocketome protein-protein interactions (Fabien Mareuil) |
| T2.13 |
PGxCorpus and PGxLOD, two shared resources for knowledge management in pharmacogenomics (Adrien Coulet) |
| T2.15 |
Expanding FORVM Knowledge graph with link suggestion for under-studied compounds (Clément Frainay) |
| T3: Evolution, phylogeny & comparative genomics |
|---|
Wed. 6th |
| T3.2 |
MPS Sampling : a novel method allowing the reliable selection of representative genomes to infer large-scale phylogenies (Coudert Rémi-Vinh) |
| T3.4 |
MicroScope: a web platform for microbial genome annotation through pangenomic and metabolic analysis (Calteau Alexandra) |
| T3.6 |
Yeast recombination specificity impact on demography inference (Louis Ollivier) |
| T3.8 |
Caractérisation du contenu en gènes de l’espèce bactérienne Coxiella burnetii issue de différentes lignées en Europe (Aminah Keliet) |
| T3.10 |
Evolution is not uniform along coding sequences (Raphaël Bricout) |
| T3.12 |
Characterization of the genomic diversity of S. Typhimurium and its monophasic variant in France in pig herds (Madeleine De Sousa Violante) |
| T3.14 |
Bacterial J-Domain Proteins and Partners Identification and Classification (Roland Barriot) |
| T3.16 |
MacSyFinder v2: An improved search engine to model and identify molecular systems in genomes (Bertrand Néron) |
| T3.18 |
Evolution of Tandemly Arrayed Genes in Rosaceae (Landès) |
| T3.20 |
Telomere-to-telomere genome assembly of the phytoparasitic nematode and virus vector Xiphinema index (Robbe-Sermesant ; Péré) |
| T3.22 |
The effect of chromosomal rearrangements on base composition evolution in mammals (Yves Clément) |
| T3.24 |
DNA methylation patterns of transcription factor binding regions characterize their functional and evolutionary contexts (Martina Rimoldi) |
| T3.26 |
Multispecies comparison of fruit development through mRNA quantification analysis (Chloé Beaumont) |
| T3.28 |
Impact of sequencing platforms on cgMLST and wgSNP analyses on species of Listeria and Salmonella (Yao Amouzou) |
| T3.30 |
Fast Construction and Extension of Gene Families (Aurélie Maurin) |
| T3.32 |
Screening of the natural two-component systems repertoire to establish guidelines for the construction of synthetic chimeras (Emilie Cottard) |
Thu. 7th |
| T3.1 |
Evaluation of word-based alignment-free methods for yeast genome comparison and taxonomy (Emmanuel Buse Falay) |
| T3.3 |
Alternative splicing modulates the number and composition of similar exonic regions (Hugues Richard) |
| T3.5 |
Comparative study of protein aggregation propensity and mutation tolerance between naked mole-rat and mouse (Savandara Besse) |
| T3.7 |
Wood-decomposing fungi through the lens of genomes comparison (Annie Lebreton) |
| T3.9 |
The genomic basis of the Streptococcus thermophilus health-promoting properties (Aurélie Nicolas - Emeline Roux) |
| T3.11 |
Most of the genetic diversity of the Wolbachia infecting Culex pipiens lies in the prophage regions (Camille Mestre) |
| T3.13 |
Deciphering the distribution of quinone biosynthetic pathways across Proteobacteria (Sophie-Carole Chobert) |
| T3.15 |
Identification of conserved Regulatory Sequences in Ray-finned Fishes (Jeanne Bauduin) |
| T3.17 |
Structural variation involving transposable elements associated with grain width in cultivated rice (Carpentier) |
| T3.19 |
Alpha-solenoid proteins for the regulation of organellar gene expression: evolutionary history and conformation studies in photosynthetic model species (Céline Cattelin) |
| T3.21 |
Gene orthology detection for Long Non Coding RNA (LncRNA) (Sandrine Lagarrigue) |
| T3.23 |
Exploration and modeling the evolution of metabolic networks in fungi (Vahiniaina Andriamanga) |
| T3.25 |
Evolution of promoter-centered 3D genome architecture across mammalian tissues (Martina Rimoldi) |
| T3.27 |
MockVirus: expanding viral phylogenetic trees by protein sequence simulation (Julia Kende) |
| T3.29 |
Automatization and optimization of TEFLoN, an accurate tool for detecting insertions of transposable elements (Corentin Marco) |
| T3.31 |
Caulifinder : a pipeline for automatic detection and annotation of endogenous viral sequences of Caulimoviridae (Helena Vassilieff) |
| T4: Functional and integrative genomics |
|---|
Wed. 6th |
| T4.2 |
A comprehensive map of preferentially located motifs reveals novel proximal cis-regulatory elements in plants (Julien Rozière) |
| T4.4 |
MYC deficiency impairs the development of effector/memory T lymphocytes (Nozais Mathis) |
| T4.6 |
Impact of genomic variation on CTCF binding and 3D genome organization in breast cancer cells (Julie Segueni) |
| T4.8 |
Research and development at the I2BC Next-Generation Sequencing Facility: an overview (Delphine Naquin) |
| T4.10 |
Automated identification of a cancer patient treatment: from sequencing to treatment prioritization (Nicolas Soirat) |
| T4.12 |
Rattus norvegicus reference genome evaluation for hippocampus RNA-seq data analysis: a glimpse into spatial transcriptomics (Christophe Le Priol) |
| T4.14 |
PDXploR, a transcriptomic comparison methodology for PDX models and matched Patient samples, a case study on osteosarcoma. (Robin Droit) |
| T4.16 |
Chromatin meta-profiling of healthy and cancer cells using publicly available datasets (Meije Mathé) |
| T4.18 |
Multi-omics and multi-tissues data to improve the understanding of heat stress adaptation mechanisms (Guilhem Huau) |
| T4.20 |
Investigating the resistance to mIDH inhibitors in Acute Myeloid Leukemia integrating multiple regulatory layers (Alexis Hucteau) |
| T4.22 |
Computational deconvolution of transcriptomics data reveals immune cell landscape of inflammatory infiltrates in giant cell arteritis (Michal Zulcinski) |
| T4.24 |
Interaction dynamics comparison of the models of Omicron BA.1 and BA.2 variants of SARS-CoV-2 Spike RBD in complex with human ACE2 through MD simulations and MM-PBSA calculations (Audrey Deyawe Kongmeneck) |
| T4.26 |
Identification of molecular subtypes from Triple-negative breast cancer tumors (Kévin Merchadou) |
| T4.28 |
AskoR, an R package for easy RNA-Seq data analysis illustrated by the analysis of plant/pathogen/microbiote interactions (Kévin Gazengel) |
| T4.30 |
Pipeline d’identification extensive de Variations Structurales dans du reséquençage nanopore de génome complet pour l’identification de mutations dans des maladies rares (Seydi Thimbo) |
| T4.32 |
scRNA-seq with Nanopore sequencing: benchmark of approaches based on hybrid sequencing (Hamraoui Ali) |
Thu. 7th |
| T4.1 |
Extensive Characterisation of Mitochondrial Genomes in Chemically Induced Mouse Liver Tumours (Maëlle Daunesse) |
| T4.3 |
Long reads RNA sequencing analysis with Oxford Nanopore Technologies: comparison of different library protocols and bioinformatics processing (Asmae Bachr) |
| T4.5 |
Analysis of single-cell RNA-seq human PBMC datasets (Eric Bonnet) |
| T4.7 |
GenomiqueENS, the IBENS Genomics core facility (Laurent Jourdren) |
| T4.9 |
Predicting clinical response to immunotherapy in melanoma (Matthieu Genais) |
| T4.11 |
RiboMethSeq platform at CRCL/CLB to profile ribosomal RNA 2’O-ribose methylation (Théo Combe) |
| T4.13 |
TnSeek : analysing Tn-seq data for multiple conditions and multiple species (Areski Flissi) |
| T4.15 |
Prediction of pediatric cancer patient progression with non-invasive procedures: Pipeline to automatize liquid biopsy analysis (Baptiste Audinot) |
| T4.17 |
Single-cell deconvolution model predictive of patient survival in clear cell Renal Cell Carcinoma (ccRCC) (Gwendoline Lecuyer) |
| T4.19 |
Single-cell ATAC-seq integration highlight epigenetics remodeling within HSC quiescence signaling. (Alexandre Pelletier) |
| T4.21 |
Detection of nucleotide repeat expansions by exome sequencing of Parkinson's disease patients using ExpansionHunter (Fanny Casse) |
| T4.23 |
Integration analysis for AAV-based gene therapy vectors with linked-read sequencing (Mallaury Vie) |
| T4.25 |
Atlas and biological significance of transcribed non-coding regions of the human genome (Pierre De Langen) |
| T4.27 |
QC impact on rare variant association results based on whole exome sequencing data (Raphael Blanchet) |
| T4.29 |
ReMap 2022: a database of Human, Mouse, Drosophila and Arabidopsis regulatory regions from an integrative analysis of DNA-binding sequencing experiments (Hammal Fayrouz) |
| T4.31 |
Harnessing gene expression profiling to infer the activation states of dendritic cell types, their dynamical relationships and their molecular regulation (Ammar Sabir Cheema) |
| T5: Metagenomics, metatranscriptomics & microbial ecosystems |
|---|
Wed. 6th |
| T5.2 |
Analyse de la diversité génétique des communautés planctoniques du Pacifique Sud-Ouest influencé par l’arc volcanique des Tonga (Charlotte Berthelier) |
| T5.4 |
metagWGS: a workflow to analyse short and long HiFi metagenomic reads (Claire Hoede) |
| T5.6 |
Construction of a reference genome catalog to decipher shared strains along an agrifood chain with shotgun metagenomic data (Solène Pety) |
| T5.8 |
A metapangenomic approach for the association of prokaryotic genes to given phenotypic traits (Nicinthya Pajanissamy) |
| T5.10 |
Towards omics-based distribution modelling of marine plankton associations at global scale (Marinna Gaudin) |
| T5.12 |
Analyzing and modelling functions carried by key species in minimal microbial communities (Chabname Ghassemi Nedjad) |
| T5.14 |
An eucaryote-friendly set of python scripts for multi-sample shotgun metagenomics and its associated Shiny application for microbiota exploration (Fiona Bottin) |
Thu. 7th |
| T5.1 |
PANORAMA : comparative pangenomics tools to explore interspecies diversity of microbial genomes (Jérôme Arnoux) |
| T5.3 |
FROGSFUNC: Smart integration of PICRUSt2 software into FROGS pipeline (Vincent Darbot) |
| T5.5 |
Eco-evolutionary diversity of the global ocean microbiome across plankton size fraction (Nicolas Henry) |
| T5.7 |
Full-length 16S rRNA gene MinION™ sequencing characterizing bacterial microbiota at species-level? (Valentine Gilbart) |
| T5.9 |
Deciphering molecular mechanisms governing malignant transformation in Neurofibromatosis Type 1 (NF1) from single cell transcriptomic (Audrey Onfroy) |
| T5.11 |
Characterising competition and cooperation potentials in microbial communities using discrete models of metabolism (Lecomte Maxime) |
| T5.13 |
Construction of ASVs networks to monitor the temporal dynamics of bacterial communities - Application to public datasets on vegetable fermentations (Romane Junker) |
| T6: Structural bioinformatics & proteomics |
|---|
Wed. 6th |
| T6.2 |
An integrative bioinformatics approach to explore the biodiversity of enzyme families (Elisée) |
| T6.4 |
Getting two birds with one stone: the Bios2cor R package for protein correlation analysis (Marie Chabbert) |
| T6.6 |
Molecular modeling of plasmodesm organization by MCTP proteins (Sujith Sritharan) |
| T6.8 |
Investigating structural and sequence determinants of regioselectivity and substrate specificity in a family of enzymes: the case study of ubiquinone biosynthesis hydroxylases (Esque) |
| T6.10 |
Towards the potentiation of selective inhibition of Mfd nanomachine (Samantha Samson) |
| T6.12 |
Modeling of NK-cell immunosurveillance in normal and pathological BM microenvironement (Berenice Schell) |
Thu. 7th |
| T6.1 |
LibProtein: a rapid and versatile annotation library for protein post-translational annotations (Hamady Ba) |
| T6.3 |
Towards molecular understanding of the UbiJ-UbiK2 protein complex by multiscale molecular modelling studies (Romain Launay) |
| T6.5 |
A new way for finding drugs target protein and discover new protein complex in CETSA experiment (Marc-Antoine Gerault) |
| T6.7 |
Reduced structural flexibility of eplet amino acids in HLA proteins (Diego Amaya) |
| T6.9 |
BioacPepFinder: Discovery of bioactive peptides from protein digestion (Isabelle Guigon) |
| T6.11 |
Structural prediction of macromolecular interactions using evolutionary information (Jessica Andreani) |
| T7: Statistics, machine learning, artificial intelligence & image analysis |
|---|
Wed. 6th |
| T7.2 |
Integrated Analyses of Large Scale RNAseq Data in Acute Myeloid Leukemia (Riedel Cédric) |
| T7.4 |
Prediction of tumor microenvironment heterogeneity in kidney cancers by cell deconvolution (Pauline Bazelle) |
| T7.6 |
Implementing a Text Mining Service Offer on the Migale Bioinformatics Platform (Mouhamadou Ba, Valentin Loux) |
| T7.8 |
Novel adversarial autoencoders to simulate human genomic data for clinical research (Callum Burnard) |
| T7.10 |
Predicting gene regulation through co-occurrence and evolutionary conservation of transcription factor binding sites (Laura Turchi) |
| T7.12 |
Dynamic genes network inference and very short time series. How repeated acoustic stimuli affect plant immunity? (Khaoula Hadj Amor) |
| T7.14 |
Using contrast to study RNA transcripts co-maturations (Benjamin Vacus) |
| T7.16 |
Robust deconvolution of transcriptomic samples using the gene covariance structure (Bastien Chassagnol) |
| T7.18 |
Feature selection in longitudinal microbiome data via the analysis of random projections (Diego Tomassi) |
| T7.20 |
Machine learning analysis on transcriptomic data reveals novel target genes of the WNT/$\beta$-catenin pathway in colorectal cancer (Andres Aravena) |
| T7.22 |
Bioinformatics integration of regulatory regions and variants in immune cells (Marie Michel) |
| T7.24 |
DetecTree, from freehand drawing to digital patient care in genomic medicine (Rafik Mankour) |
| T7.26 |
Predicting the tissue of origin from circulating DNA fragments: Biological lessons learned from a comprehensive analysis of genetic, functional and computational features to increase the accuracy of a statistical mode. (Elyas Mouhou) |
| T7.28 |
Predicting adverse drug reactions on organs using sequential neural network models. (Bryan Dafniet) |
| T7.30 |
Exploring cell morphological profile information for the de-risking of small molecules. (Fabrice Camilleri) |
| T7.32 |
Latent Dirichlet Allocation for Double Clustering (LDA-DC): Discovering patients phenotypes and cell populations within a single Bayesian framework (Elie El Hachem) |
Thu. 7th |
| T7.1 |
Investigation of neural population-based optimization (Vaitea Opuu) |
| T7.3 |
A novel method to identify and score clusters of motifs of protein sequences (CLUMPs) based on amino acids physicochemical properties. (Paola Porracciolo) |
| T7.5 |
Statistical approach for the detection of transposable element insertion bias in Drosophila melanogaster (Braham Elyes) |
| T7.7 |
Generation of a genome-wide 3D RNA profile of the catshark habenulae (Hélène Mayeur) |
| T7.9 |
Développement d’une méthode quantitative d’analyse de données RNA-Seq pour l’étude des variations in vivo des états de phosphorylation en 5’ des ARN et application chez Staphylococcus aureus (Tomas Caetano) |
| T7.11 |
Identification of epimutations in rare diseases from a single patient perspective. (Robin Grolaux) |
| T7.13 |
A general framework for classifying genomic sequences with Transformers: Application to gene annotation. (Matthias Lorthiois) |
| T7.15 |
Deep learning approaches as scoring methods for protein-protein rigid body docking. (Helene Bret) |
| T7.17 |
AOP-helpFinder : a tool for exploration of the literature to support adverse outcome pathways development (Thomas Jaylet) |
| T7.19 |
Axonal Delay Learning: from biology to computational neuroscience (Amélie Gruel) |
| T7.21 |
SciGeneX: an unsupervised method to naturally discover cell types or cell states based on patterns of co-expressed genes in single-cell RNA-sequencing data (Julie Bavais) |
| T7.23 |
Bulk RNA-Seq deconvolution for the study of hemorrhagic fever (Emeline Perthame) |
| T7.25 |
Wasserstein regularisation for multidataset PCA (Stéphane Béreux) |
| T7.27 |
Computational study of chemical-induced liver injury using high-content imaging phenotypes (Vanille Lejal) |
| T7.29 |
EnzBert : Transformer pour la caractérisation d'enzyme (Nicolas Buton) |
| T7.31 |
Automatic Empirical Segmentation of the Peritumoral Area in Lung Cancer Computed Tomography, Locating the Non-anatomical (Alexis Nolin-Lapalme) |
| T8: Systems biology & metabolomics |
|---|
Wed. 6th |
| T8.2 |
Nouvelle signature pour GSEA à partir des métabolites (Syrine Bouallegue) |
| T8.4 |
Deciphering the molecular network controlling the biology of the pig blastocyst and its cellular interactions (Adrien Dufour) |
| T8.6 |
Prioritization of Master Regulators Through Influence Maximization (Clémence Réda) |
| T8.8 |
Multi-omic integration: adding network topology to study axial spondyloarthritis (Beaudoin Annabelle) |
| T8.10 |
Logic programs to infer computational models of the human embryonic development (Mathieu Bolteau) |
| T8.12 |
What are the functions of the short open reading frame-encoded peptides in monocytes? An interactomic approach. (Sébastien A. Choteau) |
| T8.14 |
Met4J, a programmatic toolbox for graph-based analysis of metabolic networks (Ludovic Cottret) |
| T8.16 |
Improving the analysis of toxicants Mechanisms of Action with condition-specific models and network analysis (Louison) |
| T8.18 |
Logical Modeling of Dysferlinopathies (Nadine Ben Boina) |
| T8.20 |
An agent-based model of tumor-associated macrophage differentiation in chronic lymphocytic leukemia (Nina Verstraete) |
| T8.22 |
A secondary analysis of the ASSESS cohort data: the role of IFNa on genes in Primary Sjögren’s Syndrome (Diana Trutschel) |
| T8.24 |
Galaxy-SynBioCAD: tools and automated pipelines for Synthetic Biology Design and Metabolic Engineering (Thomas Duigou) |
Thu. 7th |
| T8.1 |
Chemomaps: Exploring the chimiodiversity of the living organisms (Solweig Hennechart) |
| T8.3 |
DNA methylation profiling of ATM-deficient breast tumours (Nicolas Viart) |
| T8.5 |
Modelling the dynamics of Salmonella infection in the gut at the bacterial and host levels (Muller Coralie, Wortsman Arie) |
| T8.7 |
Using machine-learning on metabolomics data to predict complex phenotypes (Sylvain Prigent) |
| T8.9 |
Metamodelling of Dynamic Flux Balance Analysis (Pablo Ugalde-Salas) |
| T8.11 |
Machine Learning classification performance on mechanistic representations of the gut microbiota built from abundance profiles (Baptiste Ruiz) |
| T8.13 |
A new Penicillium chrysogenum Genome Scale-Metabolic Network: reconciliation of previous data and focus on specialised metabolism (Delphine Nègre) |
| T8.15 |
Modélisation du métabolisme de l'adénocarcinome pancréatique (Laetitia Gibart) |
| T8.17 |
The UNTWIST project: Network analysis and performance modelling of Camelina sativa under thermal and water stress (Malo Le Boulch) |
| T8.19 |
Towards a data-driven network inference of interactions between immune and cancer cells in Chronic Lymphocytic Leukemia (Malvina Marku) |
| T8.21 |
Probing SARS-CoV-2 RNA interactome to identify post-transcriptional dysregulation associated with COVID-19 (Deeya Saha) |
| T8.23 |
A Design of Experiment strategy for GRN selection and refinement (Matteo Bouvier) |
| T8.25 |
RFLOMICS : R package and Shiny interface for Integrative analysis of omics data. (Nadia Bessoltane) |
| T9: Workflows, reproducibility & open science |
|---|
Wed. 6th |
| T9.2 |
EMERGEN-DB : The French database for SARS-CoV-2 genomic surveillance and research (Imane Messak) |
| T9.4 |
The IFB Catalogue (Hervé Ménager) |
| T9.6 |
Towards open science for deep sea sponge microbiomes (Emery Clara) |
| T9.8 |
Team efforts of the Bioinformatics and Biostatistics Hub of Institut Pasteur in response to the COVID-19 pandemic (Julien Fumey) |
| T9.10 |
Montpellier GenomiX (MGX) : next-generation sequencing and data analysis service and expertise (Xavier Mialhe) |
| T9.12 |
A collaborative methodology for MULTI-OMIC analysis (Florent Dumont) |
| T9.14 |
The Migale bioinformatics core facility (Valentin Loux) |
| T9.16 |
Single-cell Initiative of Institut Curie: presentation of technologies & bioinformatics resources (Louisa Hadj Abed) |
| T9.18 |
REPET evolutions: faster and easier (Mariene Wan) |
| T9.20 |
ePeak: from replicated chromatin profiling data to epigenomic dynamics (Rachel Legendre) |
| T9.22 |
Creation of an integrated molecular dynamics workflow on the Galaxy platform : Characterization of aquaporin pores (Agnes-Elisabeth Petit) |
| T9.24 |
Development of a pipeline integrating single-cell omic sequencing and phenotypic imaging analyses (Coline Gardou) |
| T9.26 |
PitViper: a software for comparative meta-analysis and annotation of functional screening data (Paul-Arthur Meslin) |
| T9.28 |
SCHNAPPs - Single Cell sHiNy APPlication(s) (Claudia Chica) |
| T9.30 |
Automatization of Quality Data Workflows at a Genomic Platform: the GeT-PlaGe Solution (Claire Kuchly) |
| T9.32 |
MaDMP4ls, or how to better manage bioinformatics projects with MY (Konogan Bourhy) |
Thu. 7th |
| T9.1 |
Impact environnementaux et sociaux du numérique (David Benaben) |
| T9.3 |
Omics Data Analysis Facilities in a Biomedical Research Institute (Beata Gyorgy) |
| T9.5 |
IFB training activities and resources (Khamvongsa-Charbonnier Lucie) |
| T9.7 |
The Assemblathon of the UAR 2AD, Data Acquisition and Analysis for Natural History (Marie Cariou) |
| T9.9 |
BioInformatics and Genomics platform at Institut Sophia Agrobiotech (Da Rocha Martine) |
| T9.11 |
Automated solutions for big genomic data treatment in the context of the medical diagnosis platform SeqOIA (Adrien Legendre, Thomas Rambaud) |
| T9.13 |
ABiMS: Analysis and Bioinformatics for Marine Science (Corre Erwan) |
| T9.15 |
scAN10 : A reproductible and modular Nextflow pipeline for processing 10X single cell RNAseq data (Maxime Lepetit) |
| T9.17 |
EDAM, life sciences ontology for data analysis and management. (Lucie Lamothe) |
| T9.19 |
Comparison of Stacks and a custom pipeline for RADseq analysis (Enora Geslain) |
| T9.21 |
CEA JupyterHub platform for multi-omics data analysis (Solène Mauger) |
| T9.23 |
Supports for imaging projects toward Open Science at AuBi platform (Nadia Goué) |
| T9.25 |
Assemblage hybride d'un transcriptome de novo de Solanum nigrum (Emma Corre) |
| T9.27 |
Poster UseGalaxy.fr: a Galaxy server for the French bioinformatics community (Anthony Bretaudeau) |
| T9.29 |
The IFB Core Cluster : an open HPC resource for all biologists and the breeding ground of the IFB National Network of Computational Resources (NNCR) (Gildas Le Corguillé) |
| T9.31 |
A new bioinformatics pipeline for the analysis of hepatitis B virus transcriptome by Nanopore sequencing coupled to 5’RACE (Xavier Grand) |
| Posters of demos |
|---|
| D.1 |
Comptage massif et distribué de k-mers (Alban Mancheron) |
| D.2 |
IMPatienT: an integrated web application to digitize, process and explore multimodal patient data (Corentin Meyer) |
| D.3 |
FAIR-Checker: Checking and Inspecting metadata for FAIR bioinformatics resources. (Thomas Rosnet) |
| D.4 |
Openlink, a data management dashboard for research teams (Laurent Bouri) |
| D.5 |
Phylogenomics.fr : a user-friendly web interface to phylogenomic tools and reconciliation workflow (Adrien Limouzin-Lamothe) |
| D.6 |
RiboTaxa: Combined approaches for taxonomic resolution down to the species level from metagenomics data revealing novelties (Oshma Chakoory) |
| D.7 |
ASTERICS: A Tool for the ExploRation and Integration of omiCS data (Céline Noirot) |
| D.8 |
Fantasio : Identifying rare recessive variants involved in multifactorial traits (Margot Derouin) |
| D.9 |
Snakemake RNAseq workflow including repeat expression analysis (Magali Hennion) |
| Posters of collaborative networks, working groups or associations |
|---|
| N.1 |
Expé-1point5 : une expérimentation nationale unique pour faciliter et étudier la transition des labos de recherche vers une réduction de leurs émissions de gaz à effet de serre (Schbath Sophie) |
| N.2 |
KATY european consortium: supporting the AI revolution in precision oncology (Florian Jeanneret) |
| N.3 |
ABRomics - a digital platform on antimicrobial resistance to store, integrate, analyze and share multi-omics data (Pierre Marin) |
| N.4 |
Montpellier Omics Days: An annual bioinformatics and biostatistics conference organised by Bioinformatics students (Yascim Kamel) |
| N.5 |
Green-BIM: a study to make young bioinformaticians aware of the carbon footprint of bioinformatics (Matthias Lorthiois & Hélène Dauchel) |
| N.7 |
JeBiF - Association for the Young Bioinformaticians of France (Sarah Guinchard) |
|
 Loading...
Loading...